African Trypanosome Research Published
April 1, 2024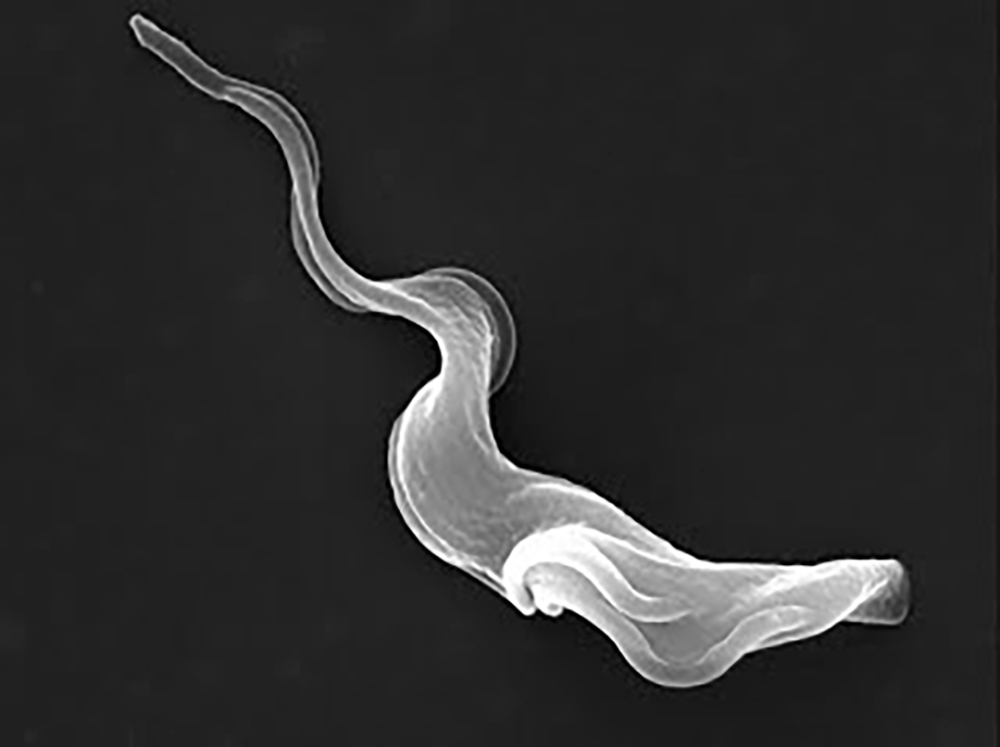
For about 35 million years, the tsetse fly and Trypanosoma brucei, a eukaryotic protozoan parasite, have been phylogenetically linked, as the parasite has adapted its surface protein to be able to live inside the fly’s midgut. However, when transmitted by the fly to a mammalian host (i.e., humans and hoofed animals), T. brucei evades the mammalian immune response by repeatedly changing its surface protein. This allows it to infect the host with the fatal African trypanosomiasis (sleeping sickness). The disease imposes a severe human and economic burden in sub-Saharan Africa where the disease is endemic. Treatments for the disease are improving, but still limited.
Enter Harvey Mudd College biology professor Danae Schulz and her students, who hope their research about how this parasite adapts to its two different hosts could lead to the design of effective new strategies for treating the disease.
The Schulz lab focuses on understanding how gene regulatory proteins called bromodomain proteins interact with the DNA as parasites transition from the blood to the insect. Over the last year, research on the topic by Schulz and several former students has been published in two scientific journals. Here is a summary of the work.
“Synthesis and characterization of I-BET151 derivatives for use in identifying protein targets in the African trypanosome,” published in Current Research in Chemical Biology, is authored by Becca Blyn ’22, Gracyn Buenconsejo ’22, Eric Tang POM ’21, Harvey Mudd College research technologist Melvin Hodanu, Schulz and students from The Wistar Institute and Thomas Jefferson University.
Trypanosoma brucei cycles between a bloodstream form in mammals and a procyclic form in the gut of its insect vector. Schulz and her students previously discovered that a human bromodomain inhibitor causes transcriptome changes that resemble the transition from the bloodstream to the procyclical form. Schulz says, “We’ve long wondered whether small-molecule bromodomain inhibitors may have other targets in African trypanosomes that might support spontaneous differentiation of bloodstream parasites to the fly stage. Using the molecules produced and analyzed in this paper, we might finally be able to answer that question.”
“Chemical Inhibition of Bromodomain Proteins in Insect-Stage African Trypanosomes Perturbs Silencing of the Variant Surface Glycoprotein Repertoire and Results in Widespread Changes in the Transcriptome,” published in the American Society for Microbiology’s journal Microbiology Spectrum, is coauthored by Jennifer Havens ’18, Lindsey Rollosson ’20, Ethan Ashby POM ’21, Schulz and Pomona College mathematics professor Johanna Hardin.
Understanding the gene-regulatory processes that facilitate transcriptome remodeling in Trypanosoma brucei may shed light on how these mechanisms evolved. “We’ve known for a long time that chemical inhibition of bromodomain proteins results in spontaneous differentiation of bloodstream parasites to the insect stage, but the effect of bromodomain inhibition on gut stage parasites was harder to study because, until recently, people didn’t know all of the genes involved in moving from the gut stage to the salivary gland stage in the fly,” Schulz says.
This paper reports that gene expression is also affected by chemical bromodomain inhibition in insect-stage parasites but that the genes affected differ depending on life cycle stage. Because trypanosomes diverged early from model eukaryotes, an understanding of how trypanosomes regulate gene expression may lend insight into how gene-regulatory mechanisms evolved. This could also be leveraged to generate new therapeutic strategies.
“With the publication of RNA-seq datasets characterizing changes in transcript levels of the relevant genes, we were finally able to analyze whether chemical inhibition of bromodomain proteins in gut stages pushes parasites toward the salivary gland stage,” Schulz says.
